EnviStor
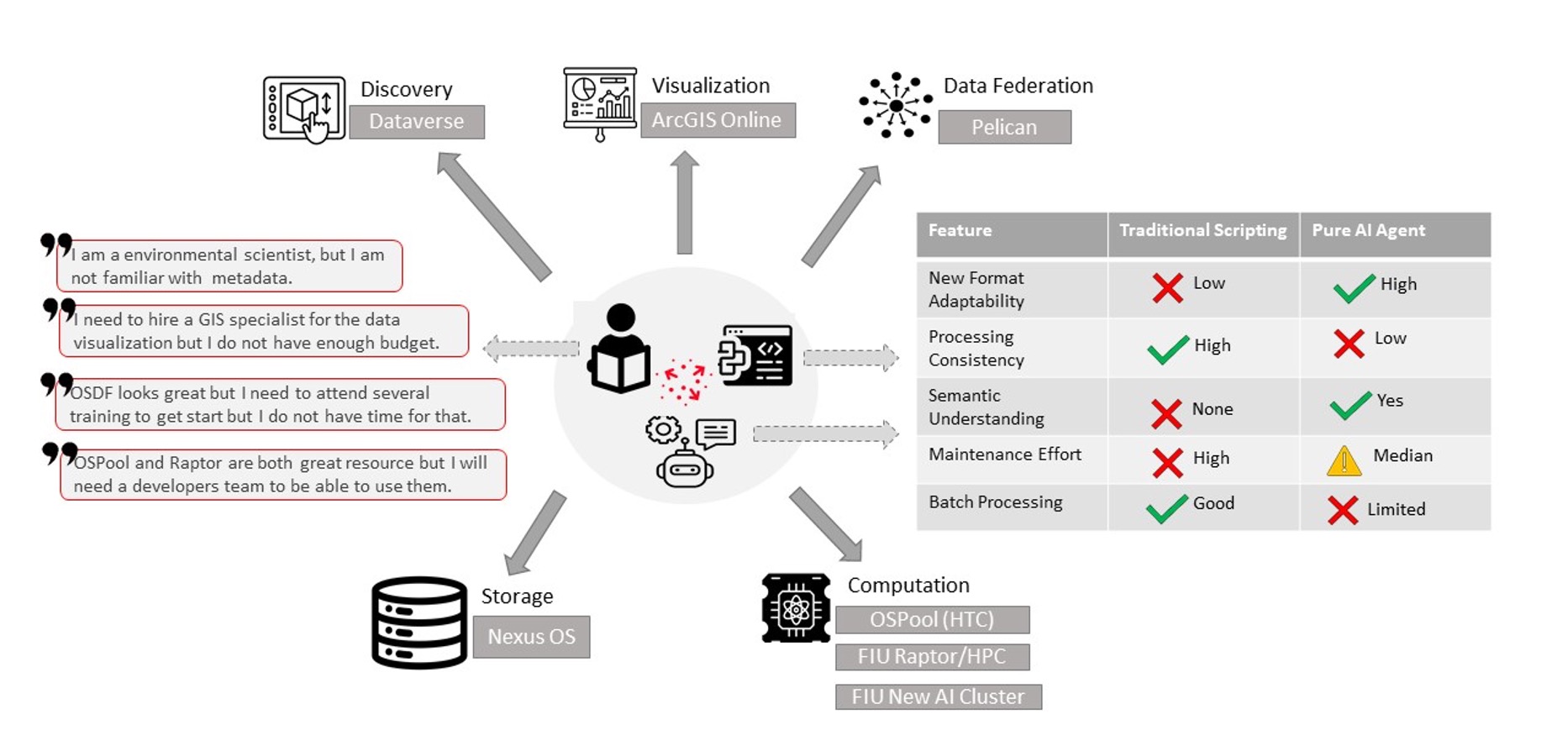
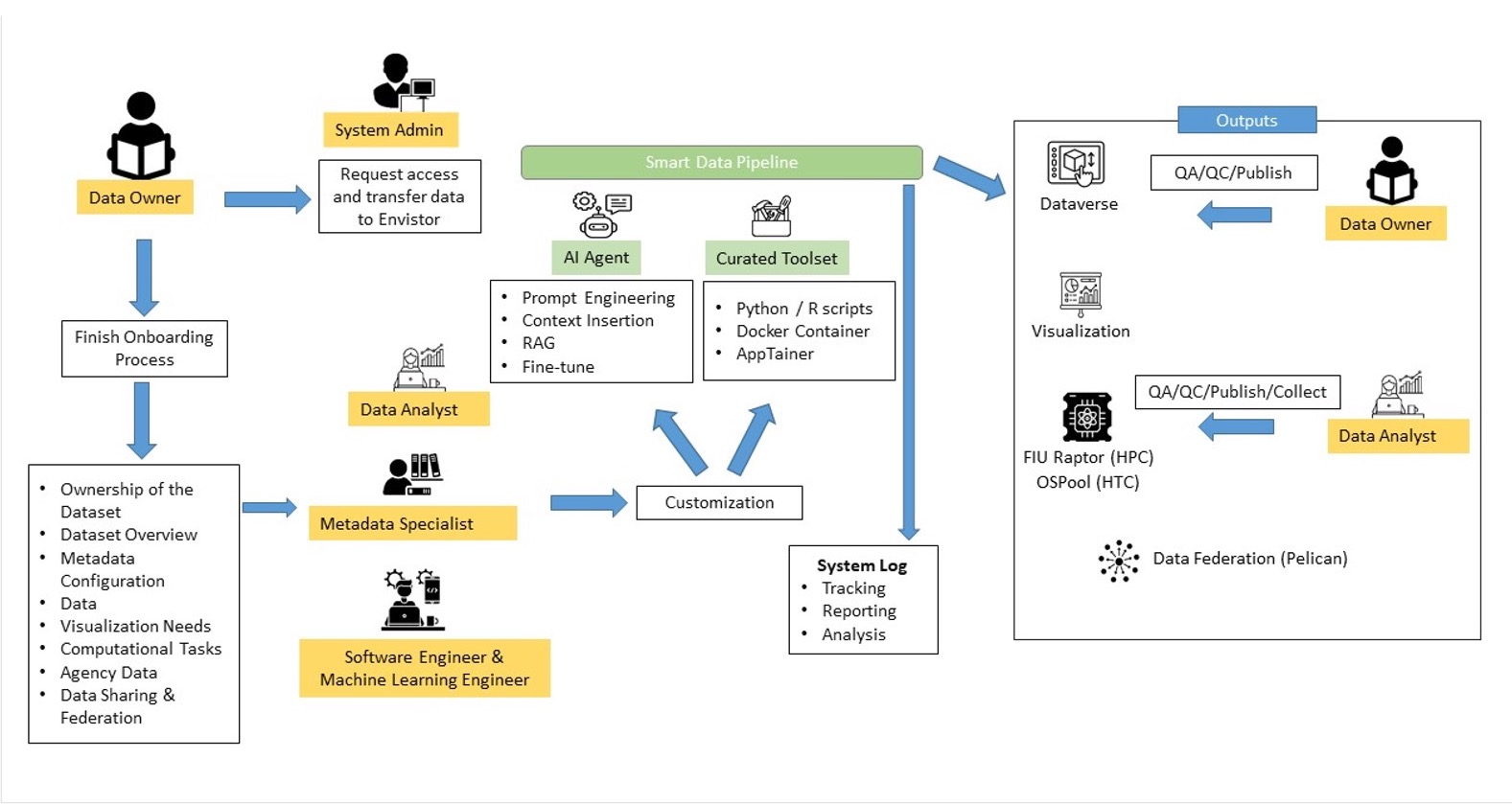
The NSF funded EnviStor is a smart data infrastructure that integrates storage, computation, metadata, and visualization to streamline research data discovery, processing, and sharing. The core components are the Harvard-developed ‘Dataverse’ , Nexus OS Ceph-backed 2.7 PB storage, ESRI’s ArcGIS, Pelican federated data sharing, OSPool (HTC) high performance computational platform. Furthermore, Envistor implements a Smart Data Pipeline to assist in data exploration, cataloging, cleaning, metadata creation, and visualization.
About
Overview
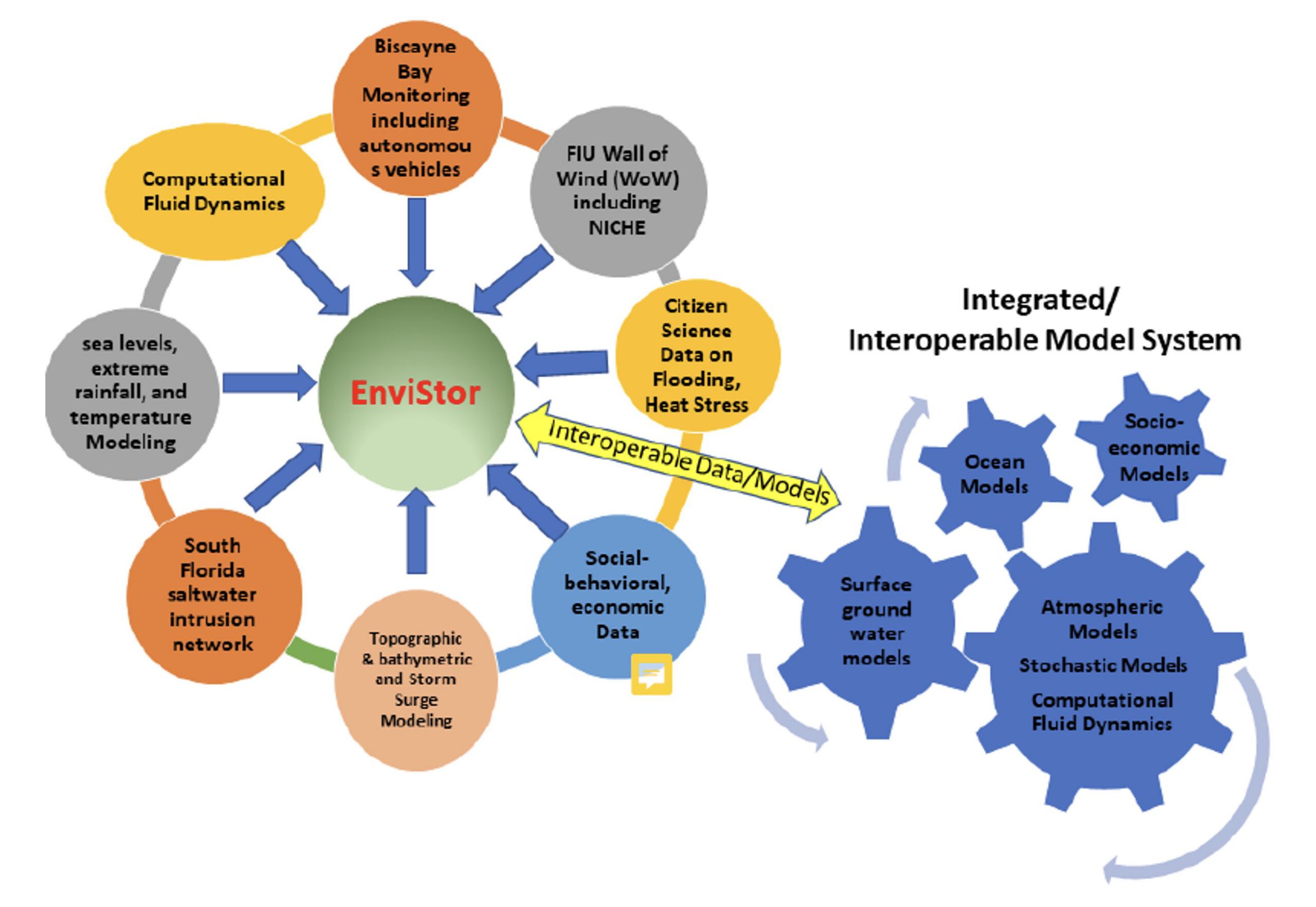
Envistor is a NSF funded data repository with 2.7 PB computing infrastructure for Supporting Collaborative Interdisciplinary Research on South Florida’s Built and Natural Environments. This repository will expand upon the Libraries’ existing research data management framework built on Harvard-developed ‘Dataverse’ (see also https://rdm.fiu.edu) and incorporate the data visualization and processing capabilities of the GIS Center, such as the geo-data. Furthermore, Envistor implements a Smart Data Pipeline to assist in data exploration, cataloging, cleaning, and metadata creation.
Currently, the project team is working on several main areas of climate and environmental data management, streaming, and visualization:
- Real-Time South Florida Saltwater Intrusion Buoy Data;
- Time Series Climate Data;
- Videos from Underwater Autonomous Vehicles;
- Wall of Wind NICHE Data;
- Time Series Remotely Sensed Imagery Data; 6) Storm surge and flood simulation data and models.
The EnviStor system connects to the NSF federated data computing system “Pelican”, as well as FIU’s Reconfigurable Advanced Platform for Transdisciplinary Open Research (RAPTOR) to support nationwide Open Science data federation. FIU Libraries GIS Center and DCC will play an essential role in configuring and implementing the EnviStor IT framework, as well as in AI-assisted data management, dashboard creation, metadata management, and web application development for the user interface of EnviStor.
Workflow and Contact
This Standard Operating Procedure Document sets forth procedures for the flow of datasets (geospatial and non-geospatial) through the FIU Envistor infrastructure. It contains workflows for adding/publishing datasets/items by utilizing Envistor Smart Data Pipeline, which can automate the following tasks:
- Automated metadata generation and publishing datasets in Dataverse;
- Data visualization Applications, e.g. ArcGIS Online (Optional and fee-based);
- Advance Computational Tasks, e.g. Data/Image Processing (Optional and fee-based);
- Connect to OSDF/Pelican federation (Optional and fee-based).
Contact
As a first step, make sure that you and your team have authenticated access to upload data to Envistor (download instructions: EnviStor_Rclone_Configuration_v1 1 1 1.pdf), and contact Anthony Bellantuono (ajbellan@fiu.edu) and Ernesto Rivas (erivas@fiu.edu).
For batch data upload, please prepare your data information in the spreadsheet for the Envistor Agency/Third Party datasets or Envistor FIU research data metadata elements datasheets. If you have any questions relating to the metadata generation within Dataverse, please contact Sonia Santana Arroyo or Rebecca Bakke at rdm@fiu.edu.
For questions relating to Smart Data Pipeline workflow, please contact Boyuan Guan (bguan@fiu.edu).
For Pelican/OSPool access, please contact Dr. Leonardo Bobadilla (bobadilla@cs.fiu.edu).
Collaborators
FIU’s Knight Foundation School of Computer and Information Sciences
FIU Libraries GIS Center and Digital Collection Center
Division of IT
Institute of Environment
Sea Level Rise Solution Center
Institute of Extreme Events
Tech Support Teams
Smart Data Pipeline team: Boyuan Guan (bguan@fiu.edu), Amandha Wingert Barok (awingert@fiu.edu), Wencong Cui (wecui@fiu.edu), Pratik Poudel (ppoudel@fiu.edu)
EnviStor IT team: Anthony Bellantuono (ajbellan@fiu.edu), Anu Chirinos (anu@fiu.edu), Ernesto Rivas (erivas@fiu.edu)
Metadata support: Sonia Santana Arroyo (sonsanta@fiu.edu), Rebecca Bakke (rbakker@fiu.edu)
Website: Julian Gottlieb (gottliej@fiu.edu)
What is the FIU EnviStor, and which FIU research units are involved?
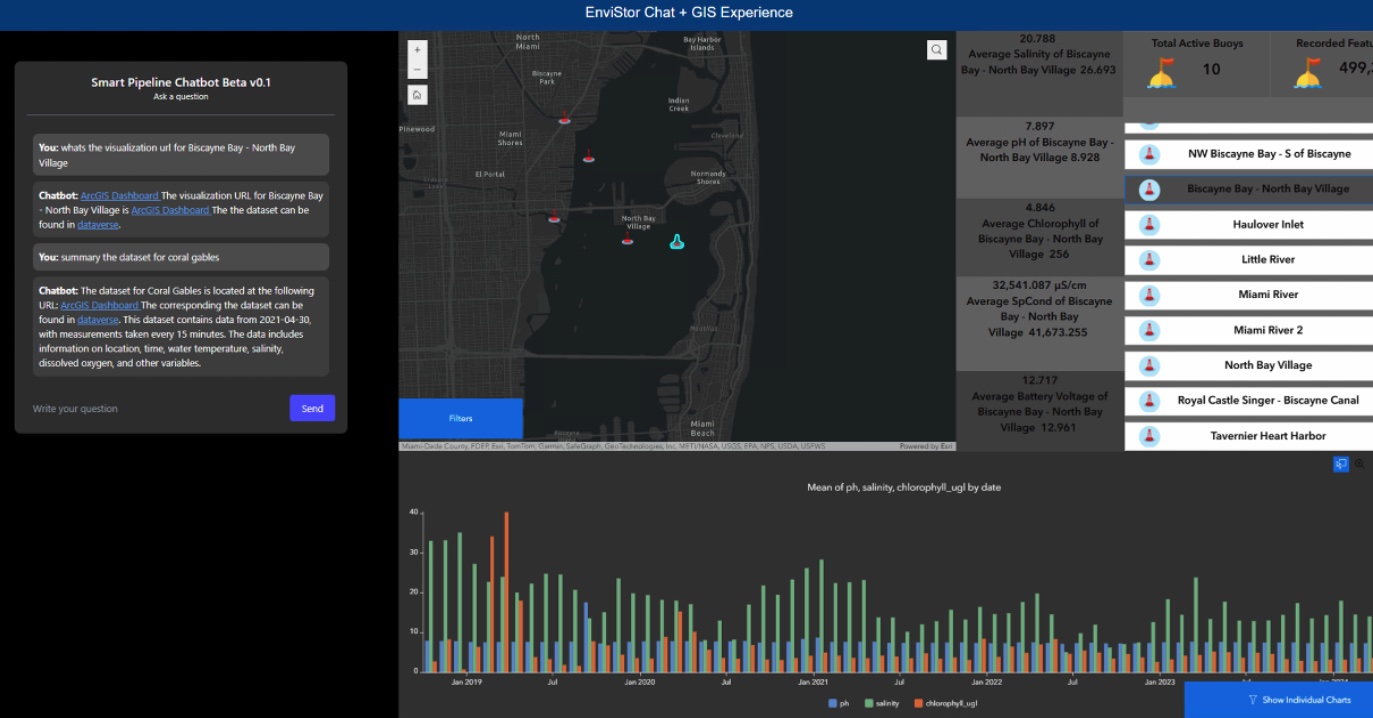
Envistor is a smart data infrastructure that integrates storage, computation, metadata, and visualization to streamline research data discovery, processing, and sharing for environmental datasets of the South Florida Region. Examples of datasets are Bouy data, climate (rainfall, temperature) data, elevation, storm surge, aerial imageries, autonomous underwater vehicle captured datasets, as well as Wall of Wind video files, etc. The NSF funded Envistor project is a collaborative effort among FIU’s DoIT, Knight Foundation School of Computing and Information Sciences, FIU Libraries GIS/Digital Collection Centers, Institute of Environment, Sea Level Solution Center, Extreme Event Center, School of Engineering.
What is the FIU Envistor Dataverse, and why do we need metadata?
Dataverse is an open-source web application designed to store, share, cite, and publish research data. It provides a platform where researchers can manage and share datasets in a structured, secure, and accessible way. Dataverse is often used by academic institutions, research organizations, and data repositories to promote transparency, reproducibility, and collaboration in research.
Key Features of Dataverse:
- Dataset versioning and file-level access control
- Integration with DOIs (Digital Object Identifiers) for data citation
- Metadata support for discovery and reuse
- Data curation tools and analytics integration
- Customizable metadata schemas for different disciplines
FIU Dataverse serves as a data catalog for discovery of datasets, supporting documents, scripts, codes tools and models in Envistor. We aim to manage two different types of datasets – FIU researcher generated, collected, processed, or value-added data, and data downloaded from government agencies or other third parties. Every item in the FIU Envistor should contain high-quality metadata. Including high-quality metadata while publishing/adding items to the Dataverse is crucial in ensuring the smooth transfer of these items to the Pelican, and ultimately, in making discovery of these datasets possible by the end users within the federated computing network.
For more information about viewing and editing metadata in Dataverse manually: User Guide — Dataverse.org.
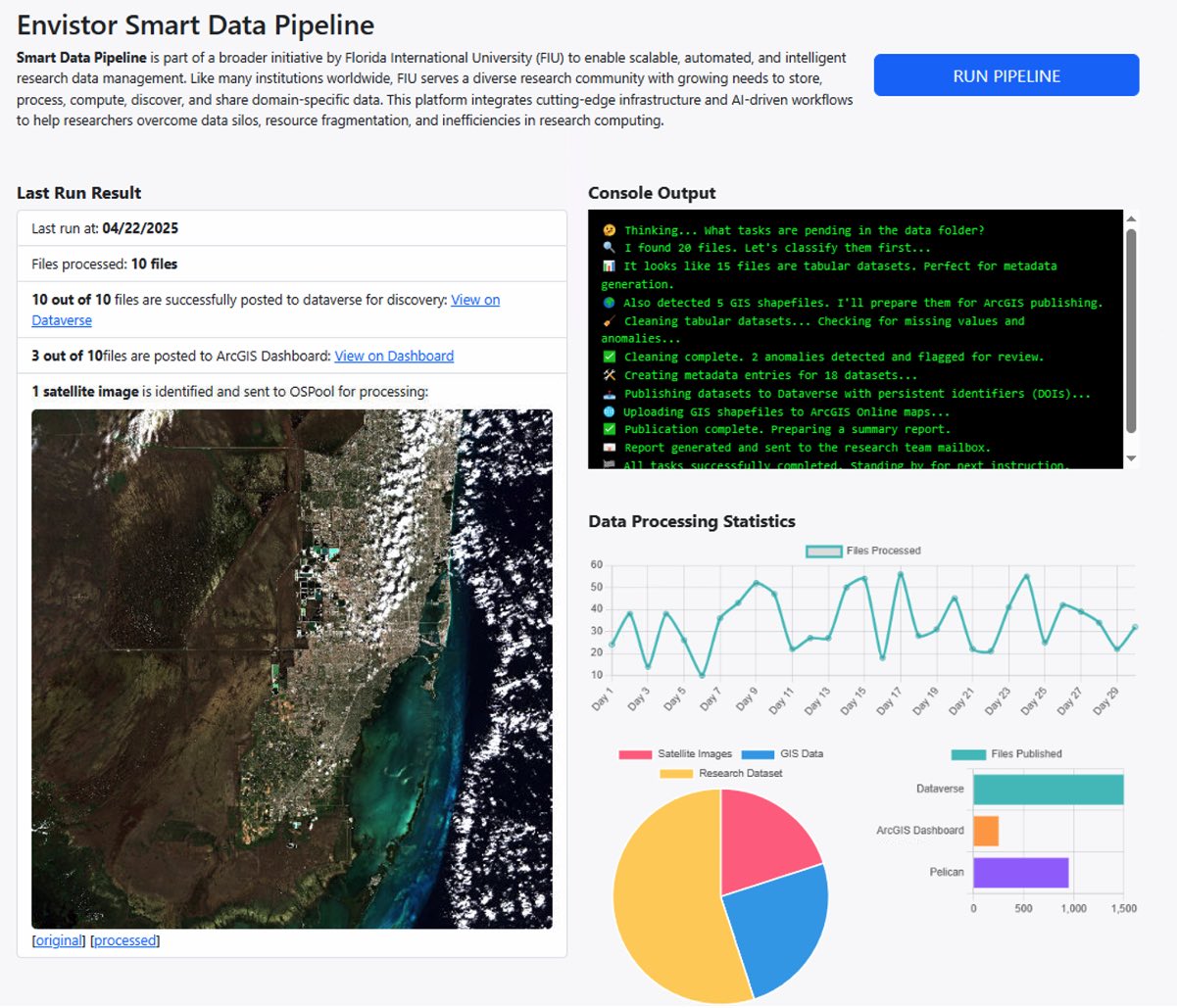
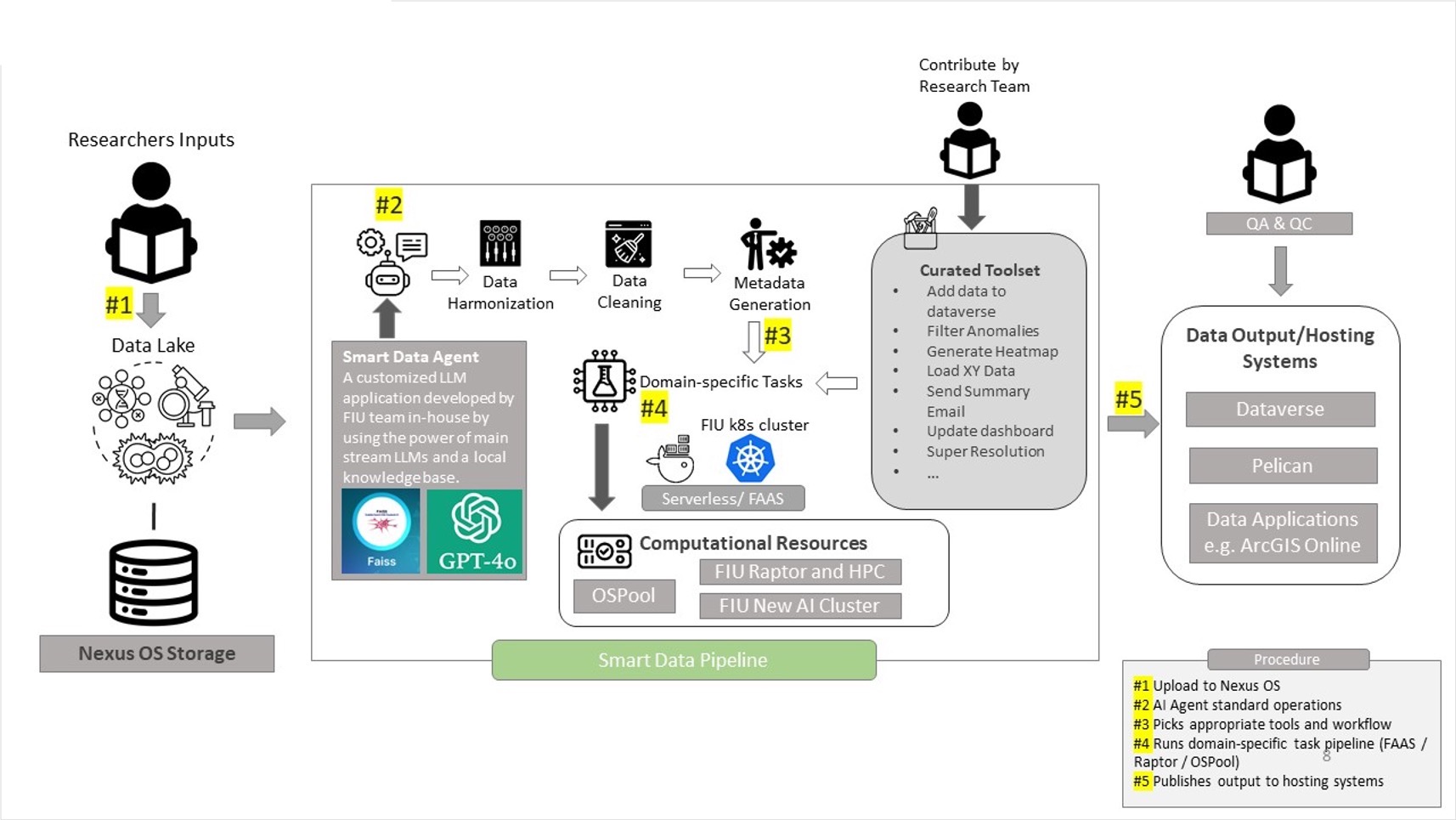
What is a Smart Data Pipeline and how does it work?
Smart Data Pipeline is part of a broader initiative by Florida International University (FIU) to enable scalable, automated, and intelligent research data management. Like many institutions worldwide, FIU serves a diverse research community with growing needs to store, process, compute, discover, and share domain-specific data. This platform integrates cutting-edge infrastructure and AI-driven workflows to help researchers overcome data silos, resource fragmentation, and inefficiencies in research computing.
What is OSPool/Raptor, and how does it work?
The Open Science Pool (OSPool) is a distributed high-throughput computing (dHTC) platform operated under the Open Science Grid (OSG). It federates compute resources from multiple institutions and provides a scalable, flexible, and cost-effective environment to run large numbers of computational tasks in parallel.
Key Features:
- Massive scale: Ideal for workloads that consist of many independent jobs (e.g., data preprocessing, simulations, model evaluations).
- Data-aware scheduling: Paired with OSDF (Open Science Data Federation), OSPool can access data in-place from federated storage without unnecessary data movement.
- Federated access: Supports workflows that span multiple campuses and HPC sites.
Role in EnviStor:
In the EnviStor framework, OSPool is leveraged to perform large-scale computation on federated datasets, especially from Pelican-based storage systems. When data is stored and registered through OSDF/Pelican, users can run jobs on it via OSPool without needing to replicate the datasets across systems. This dramatically increases the efficiency of processing large environmental datasets like buoy time series, sensor data, or remote sensing outputs (SmartDataPipeline4BouyD…).
RAPTOR (Reconfigurable Advanced Platform for Transdisciplinary Open Research) is an NSF-funded high-performance computing (HPC) cluster hosted at FIU. It is designed to support data-intensive and compute-heavy research by offering configurable compute environments for diverse workflows.
Key Features:
- Integrated with OSG for federated job submission.
- Provides low-latency and high-throughput compute nodes.
- Configured to accept jobs via OSG Connect and HTCondor.
Role in EnviStor:
In the EnviStor ecosystem, RAPTOR serves as a local high-performance compute backend. It works in conjunction with OSPool and the broader FIU compute cluster to:
- Process computational workloads submitted via OSG Connect.
- Run AI models, simulations, or post-processing steps on structured data stored in EnviStor’s /scratch space.
- Accept job submissions managed by an OSG controller, which routes them to RAPTOR via secure login nodes.
What is Pelican, and why do we choose it?
Pelican is a data federation platform developed by the Partnership to Advance Throughput Computing (PATh) to support the Open Science Data Federation (OSDF). It allows institutions to expose datasets stored in POSIX filesystems, S3 buckets, or HTTP servers as part of a distributed, federated storage network.
We selected Pelican for the following reasons:
- Federated Data Sharing
Pelican enables distributed data access without centralizing storage. Researchers can publish datasets from their local storage systems while making them accessible through a global, persistent pelican:// URL. - Compute-Aware Federation
Datasets registered with Pelican can be directly accessed by compute platforms like OSPool, enabling data-local computation. This reduces data movement overhead and improves performance for large-scale workflows. - Scalability and Flexibility
Pelican supports a wide range of storage backends (POSIX, S3, HTTP), making it compatible with FIU’s existing infrastructure (e.g., Ceph, OSNexus). It integrates well with our storage pools (/data1 and /data2), allowing flexible data organization. - FAIR Support via Metadata Linking
While Pelican itself has minimal metadata capabilities, it integrates seamlessly with Dataverse, which manages rich metadata. We use Dataverse for metadata discoverability and DOI assignment, and link it to data stored in Pelican, ensuring compliance with the FAIR (Findable, Accessible, Interoperable, Reusable). - Open-Source and Community-Aligned
Pelican is part of an actively maintained, open-source ecosystem that aligns with EnviStor’s mission to support inter-institutional research and shared infrastructure.
What is the recommended file naming convention for Envistor?
When publishing datasets to FIU Envistor, we recommend but not require follow the naming convention outlined in the table below.
This file naming structure captures file type, keywords, geo name, and year of the datasets. For datasets downloaded from the agencies, you have a choice of keep the original file naming conventions and structure, as they may have an existing file naming convention.
|
OBJECT TYPE |
PREFIX |
Example |
|
Vector |
v_ |
v_rivers_South Florida_yyyy |
|
Raster (Imagery or tile-based raster) |
r_ |
r_DoQQ_Bounding Coordinates_yyyy |
|
Table/Spreadsheet |
t_ |
t_Waterquality_Biscayne_yyyy |
|
Notebook |
n_ |
n_HourlyRainFall_Extraction_Georegion_yyyy |
|
Script (Py or R) |
s_ |
s_GeoStatistical Processing_R; or s_classifier_Py |
|
Model |
m_ |
m_innudation bathtub |
|
Video |
V_ |
V_Keyword_yyyy |
|
Zip |
Z_ |
Z_keyword_yyyy |
What data file formats can Envistor Dataverse accommodate?
FIU Envistor accommodates various data formats:
- Shapefiles/feature classes (zip, or .shp)
- Spreadsheets (.csv)
- Jupyter notebooks, PY Scripts, codes (_R, _Py)
- Imagery or tile-based raster (e.g. elevation)
- Other (PDF, API, etc.)
The most used environmental and climate data formats and tools are as follow:
🔷 1. NetCDF (.nc, .nc4)
🔧 Tools / Libraries:
- Python:
- xarray – powerful, label-aware handling of NetCDF data
- netCDF4 – low-level library for reading/writing
- cfgrib – read GRIB files as if they were NetCDF
- R:
- ncdf4, raster, stars
- MATLAB:
- Built-in support via ncread, ncinfo
- Command-line:
- ncdump, ncks, ncrcat (from NCO toolkit)
- cdo (Climate Data Operators)
- GUI Tools:
- Panoply (by NASA GISS) – visualizes NetCDF
- QGIS – can import NetCDF as raster layers
🔷 2. HDF/HDF5 (.hdf, .h5)
🔧 Tools / Libraries:
- Python:
- h5py – low-level HDF5 access
- netCDF4 – supports NetCDF-4 (built on HDF5)
- xarray – indirect support
- R:
- rhdf5, hdf5r
- MATLAB:
- Built-in support with h5read
- Command-line:
- h5dump, h5ls (from HDF Group)
- NASA Software:
- HEG Tool – reprojects and mosaics MODIS HDF files
🔷 3. GeoTIFF (.tif, .tiff)
🔧 Tools / Libraries:
- Python:
- rasterio – excellent for raster I/O
- xarray + rioxarray
- GDAL (via osgeo.gdal)
- R:
- raster, terra
- GIS Tools:
- QGIS, ArcGIS, GRASS GIS
- Command-line:
- gdalinfo, gdal_translate, gdalwarp
🔷 4. GRIB (.grib, .grb, .grb2)
🔧 Tools / Libraries:
- Python:
- cfgrib (uses ecCodes backend)
- pygrib – lower-level interface
- R:
- rNOMADS (limited)
- Command-line:
- wgrib, wgrib2 – very efficient parsing
- cdo, ncl – widely used in climate modeling
- GUI:
- Panoply, QGIS (via plugin)
🔷 5. CSV (.csv)
🔧 Tools / Libraries:
- Python:
- pandas – ideal for time series and tabular data
- R:
- csv, readr::read_csv
- Excel / LibreOffice Calc
- GIS:
- Import as points in QGIS/ArcGIS if lat/lon columns exist
🔷 6. Shapefile (.shp, .shx, .dbf, etc.)
🔧 Tools / Libraries:
- Python:
- geopandas – loads shapefiles easily
- fiona (lower-level)
- R:
- sf, rgdal, sp
- GIS:
- QGIS, ArcGIS, GRASS GIS
- Command-line:
- ogrinfo, ogr2ogr (from GDAL)
🔷 7. GeoJSON (.geojson, .json)
🔧 Tools / Libraries:
- Python:
- geopandas, json, folium, leaflet
- R:
- sf, geojsonio, leaflet
- Web:
- js, Mapbox, OpenLayers
- GIS:
- QGIS, ArcGIS
🔷 8. Zarr
🔧 Tools / Libraries:
- Python:
- zarr – core library
- xarray – supports Zarr natively
- Often used with Pangeo stack (Dask, Xarray, Zarr)
- Cloud:
- Cloud-native object stores (e.g., S3, GCS)
- Works well in JupyterHub/Dask setups for scalable analysis
If I have data that I would like to release weekly/daily/hourly in batches to the public (collections of images captured that I would like to be public), would I just need to fill out the form once to denote that there will be a group of images being released and updated constantly?
That’s correct. The smart data pipeline will be handling the processing of the data and generation of the metadata. However, the spreadsheet of metadata (e.g the form) should be completed by the data owners/curators.
What would I need to do from my side, automation wise, to denote a set of images as marked for the next public release? I.e. Moving images to a specific directory within the object store, providing a prefix to their keys to denote that they should be public, etc. (this would all be from my api side).
Firstly, just move your data into the public bucket. We might need separate technical meetings if special needs are required; otherwise, we will harvest the data into Dataverse based on your responses you fill in the spreadsheet.
Would it be helpful to your EnviStor team if I created a little file that could act as basically an instructional markdown document on how to interact with the EnviStor object store if one is using Node.js with the AWS Node.js SDK? Providing instructions on how to put objects, list objects, retrieve objects, and most importantly how to handle credential management to be able to connect to the s3 object store since the docs are really confusing on that front.
For sure it will be helpful. The Envistor support team will work closely with data analysts and data owners to customize to their desired computing environment for the repository. The more knowledge or expertise the domain data analysts can provide, the better.
Could Envistor provide Code Archival via GitLab?
That’s a great suggestion, and it’s something we’ve considered during our architectural planning. However, to clarify—the Smart Data Pipeline is not intended to function as a traditional DevOps pipeline (e.g., CI/CD for code builds or deployments).
Instead, our focus is on reproducibility, portability, and data-centric automation. To support this, each data owner’s environment—including their scripts, workflows, and supporting tools — is encapsulated within a container. This ensures that:
- Teams can share and execute their work consistently across environments (e.g., from local development to OSPool and Raptor).
- We avoid dependency drift and complex setup issues.
- Processing tasks can be federated or scheduled on distributed resources.
Given this design, an internal Git repository service (e.g., GitLab) is not part of the core infrastructure. That said, we do encourage teams to maintain their own version control systems (whether GitHub, GitLab, or Gitea, we are using a private Bitbucket ourselves) for managing development and source code internally.
In the future, if we see strong demand across teams for an in-house Git service—particularly one that integrates with the Smart Data Pipeline metadata workflow—we’re open to revisiting this. For now, our goal is to support code and workflows through containerization rather than integrating full DevOps pipelines.
However, source code can also be uploaded into Dataverse but the intent here is not for code reuse or direct execution. Instead, the purpose is:
- Cataloging the computational assets tied to a dataset or research output
- Enabling discovery and transparency (e.g., letting others understand what methods or tools were used)
- Supporting citation and long-term preservation of the research process
Since the Smart Data Pipeline treats containers as the actual execution unit (with dependencies, configuration, and runtime setup baked in), the source code in Dataverse acts more like a research artifact—similar to a methods appendix in a paper.
You can think of it this way (our current setup):
- Bitbucket is for active code development and internal collaboration
- Containers are for standardized, portable execution
- Dataverse is for archiving and documenting what was used to generate a research output
This layered model ensures separation of concerns while supporting traceability and compliance with FAIR principles.